Why R?
The old way...
M to see all slides, G to go to a specific slide
R is a language that's easy to learn badly

glm(y ~ -1 + a + c + z + a:z, data = mydata, maxit = 30)
All come with key bindings. Pick one that you like.
load.r |
→ |
clean.r |
→ |
functions.r |
→ |
do.r |
# File: do.r source("functions.r") load("cleaned_data.rdata")
# My_script.r setwd("path/to/dir") # Removing any extra objects from my workspace (just in case) rm(list = ls()) # Keeps my current file clutter free source("related_r_scripts.r")
attach(mydata)
mydata$column_name
Global Environment |
Function or Namespace |
Good ✔ |
Bad ✖ |
# Vector: (single dimension, all same type) vec1 <- 1:10 class(vec1)## [1] "integer"vec2 <- letters[1:10] class(vec2)## [1] "character" # Data Frame: Each column is a vector, but adjacent vectors can hold different things # Matrix: Just like a data.frame except it's all numeric # List: (any dimension, mix and match)l1 <- list(A = data.frame(x = 1:10, y = rnorm(10)), B = 1, C = letters[1:3]) str(l1)## List of 3 $ A:'data.frame': 10 obs. of 2 variables: ..$ x: int [1:10] 1 2 3 4 5 6 7 8 9 10 ..$ y: num [1:10] 0.618 0.519 0.343 0.428 -0.885 ... $ B: num 1 $ C: chr [1:3] "a" "b" "c"
?function_name
??function_name
RSiteSearch("function_name")
StackOverflow.com/questions/tagged/r
 |
Rseek.org
 |
sessionInfo()sessionInfo()## R version 2.14.2 (2012-02-29) Platform: x86_64-apple-darwin9.8.0/x86_64 (64-bit) locale: [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8 attached base packages: [1] graphics grDevices utils datasets stats methods base other attached packages: [1] knitr_0.3 formatR_0.3-4 devtools_0.6 plyr_1.7.1 [5] reshape2_1.2.1 ggplot2_0.9.0 loaded via a namespace (and not attached): [1] codetools_0.2-8 colorspace_1.1-1 dichromat_1.2-4 [4] digest_0.5.1 evaluate_0.4.1 gdata_2.8.2 [7] grid_2.14.2 gtools_2.6.2 highlight_0.3.1 [10] MASS_7.3-17 memoise_0.1 munsell_0.3 [13] parser_0.0-14 proto_0.3-9.2 RColorBrewer_1.0-5 [16] Rcpp_0.9.10 RCurl_1.91-1 scales_0.2.0 [19] stringr_0.6 tools_2.14.2
dput() to share some datadput(head(mtcars))structure(list(mpg = c(21, 21, 22.8, 21.4, 18.7, 18.1), cyl = c(6, 6, 4, 6, 8, 6), disp = c(160, 160, 108, 258, 360, 225), hp = c(110, 110, 93, 110, 175, 105), drat = c(3.9, 3.9, 3.85, 3.08, 3.15, 2.76), wt = c(2.62, 2.875, 2.32, 3.215, 3.44, 3.46), qsec = c(16.46, 17.02, 18.61, 19.44, 17.02, 20.22), vs = c(0, 0, 1, 1, 0, 1), am = c(1, 1, 1, 0, 0, 0), gear = c(4, 4, 4, 3, 3, 3), carb = c(4, 4, 1, 1, 2, 1)), .Names = c("mpg", "cyl", "disp", "hp", "drat", "wt", "qsec", "vs", "am", "gear", "carb"), row.names = c("Mazda RX4", "Mazda RX4 Wag", "Datsun 710", "Hornet 4 Drive", "Hornet Sportabout", "Valiant"), class = "data.frame")
.rprofileoptionsoptions(max.print = 2000) options(prompt = "$ ") options(stringsAsFactors = FALSE) # Store API keys options(MendeleyKey = "My_secret_key")
See ?options for more information on settings
options() to list current settings
.rprofilelibrary(ggplot2) library(stringr) library(plyr) library(devtools)
.rprofile# A function that tells me which packages are out of date check.packages <- function() { if (!is.null(utils::old.packages())) { old_packages <- utils::old.packages() cat("Notification:", dim(old_packages)[1], "packages are out of date \n") cat(unname(old_packages[, 1]), sep = ",", "\n") } if (is.null(utils::old.packages())) { cat("All packages are current \n") } }
.rprofile.rprofile does make life convenient, remember that any code/settings stored there are not reproducible by others. # To load R without the .rprofileR -- vanilla
Thanks to Hadley for pointing this out oversight.
read.tableUse packages RODBC, RMySQLAmazon S3, Google Docs, Dropbox etc.Long-termsave(data, file = "slots.rdata")
write.table(data, file = "slots-3.csv", sep = ",", row = F)
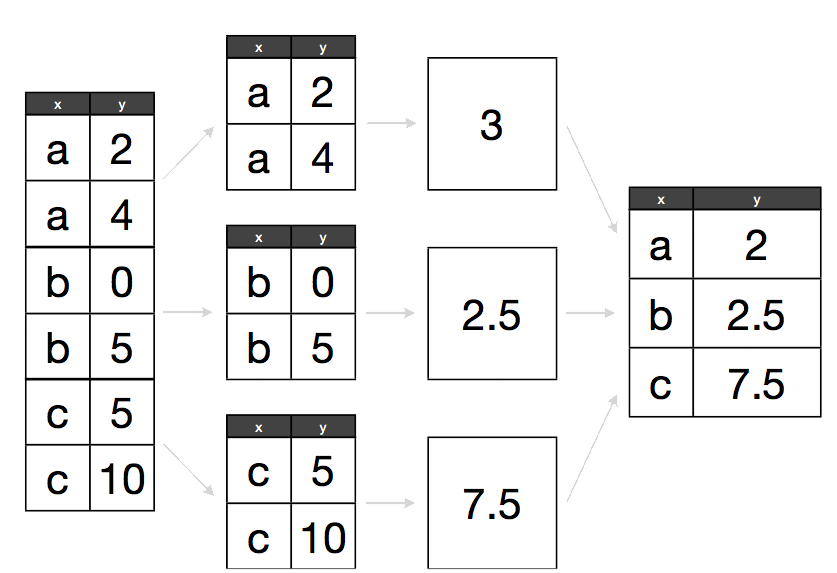
Hadley Wickham (2011). The Split-Apply-Combine Strategy for Data Analysis. JSS, 40(1), 1-29. for more details
data <- data.frame(x = c("a", "a", "b", "b", "c", "c"), y = c(2, 4, 0, 5, 5, 10))
ddply(data, .(x), summarise, y = mean(y))
result <- ddply(data, variable, summarise, n = sum(n))
result <- llply(list_name, function_name)
dcast(melted_data, temp ~ light, length)
dcast(melted_data, temp ~ light, mean)
dcast(melted_data, temp ~ light, custom_function)
test_data <- data.frame(id = 1:9, category = factor(rep(sample(letters[1:3]), 3)), treatment = rep(sample(c("control", "trt_1", "trt_2")), 3), price_index = rnorm(9) * 200, prev_yr_index = rnorm(9) * 200)
melted_data <- melt(test_data, id.vars = 1:3)
dcast(melted_data, category + treatment ~ variable, length) dcast(melted_data, category + treatment ~ variable, mean)
If you have to repeat the same 3-4 lines of code more than once, turn it into a function


# As easy as:
knit('report.rnw')
# All the syntax in this talk was generated using knitr

/
#