Data Life Cycle
source: Michener, 2006 Ecoinformatics.
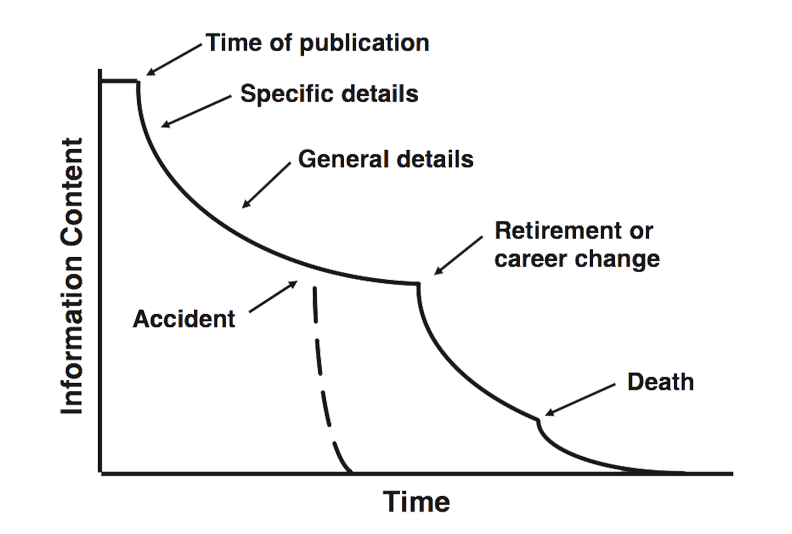
Most science is not reproducible or repeatable, even within the same lab group over time.

source: Michener, 2006 Ecoinformatics.
glm(y ~ -1 + a + c + z + a:z, data = mydata, maxit = 30)
Data repositories | ||||
 |
 |
 |
||
 |
 |
|||
 |
||||
Literature | ||||
 |
 |
|||
Metadata | ||||
 |
 |
 | ||
 |
API keys can be stored in a users.rprofile
options(MendeleyKey = "uf5daib7wyil7ag5buc") options(MendeleyPrivateKey = "faj2os5dyd7jop2fok6") options(PlosApiKey = "ef3vip9yak7od3hud4g") options(SpringerMetdataKey = "ri9hi7woc6jax4vaf8w")
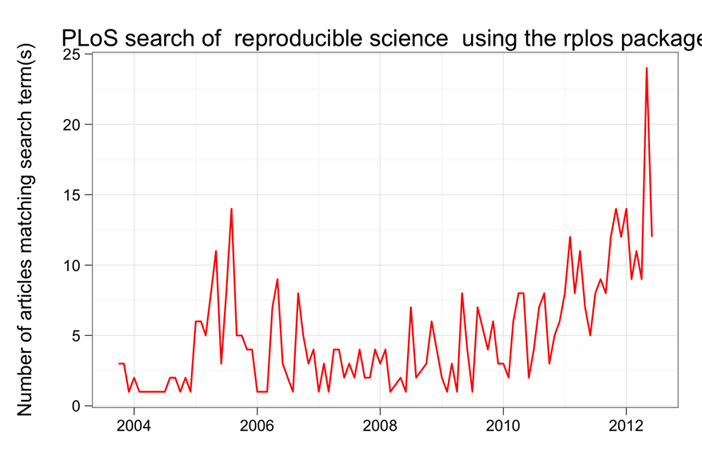
rploslibrary(rplos) plot_throughtime(list("reproducible science"), 500)
RMendeleygroupDocInfo(mc, 530031, 4344945792)
$abstract
[1] "SUMMARY: Modern biological experiments create vast amounts of data which are geographically distributed. These datasets consist of petabytes of raw data and billions of documents. Yet to the best of our knowledge, a search engine technology that searches and cross-links all different data types in life sciences does not exist.....
$authors
$authors[[1]]
forename surname
"Dominic S" "L\xfctjohann"
# ....
dryad# Get the URL for a data file dryaddat <- download_url("10255/dryad.1759") # Get a file given the URL file <- dryad_getfile(dryaddat)
raltmetGitHub(userorg = "ropensci", repo = "rmendeley")
totimp(id = "10.5061/dryad.8671")
stackexchange(ids = 16632)
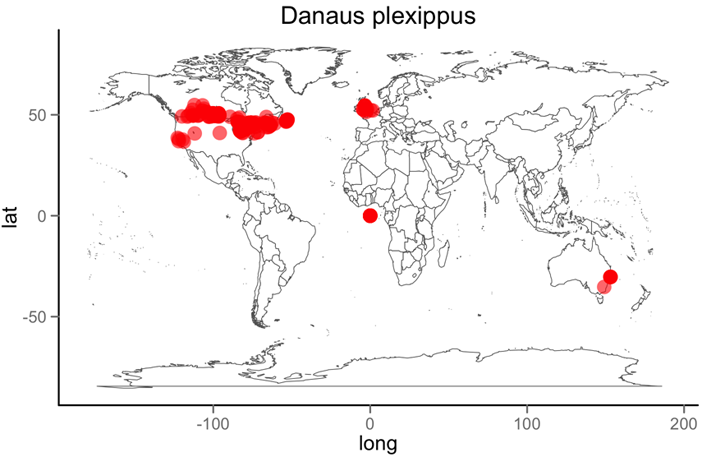
rgbifdistribution <- occurrencelist(sciname = "Danaus plexippus", coordinatestatus = TRUE, maxresults = 1000, latlongdf = TRUE)
(figshare)R directly to one's figshare account.> figshare(data) # code isn't ready yet but once it is, it will return a persistent identifier

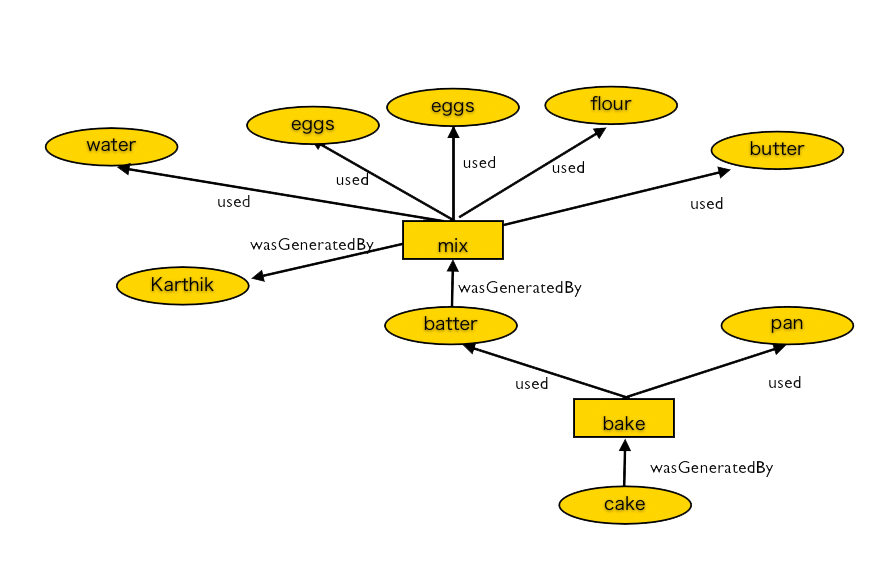
Source: Modified from original version by James Cheney, University of Edinburgh.
knitr where a user can track workflow provenance using hooks.XML to track metadata and maintain provenance traces across runsknitr + Markdown + GitHubGitHub automatically renders Markdown and even provides syntax highlighting
knitr + Markdown + GitHub = executible paper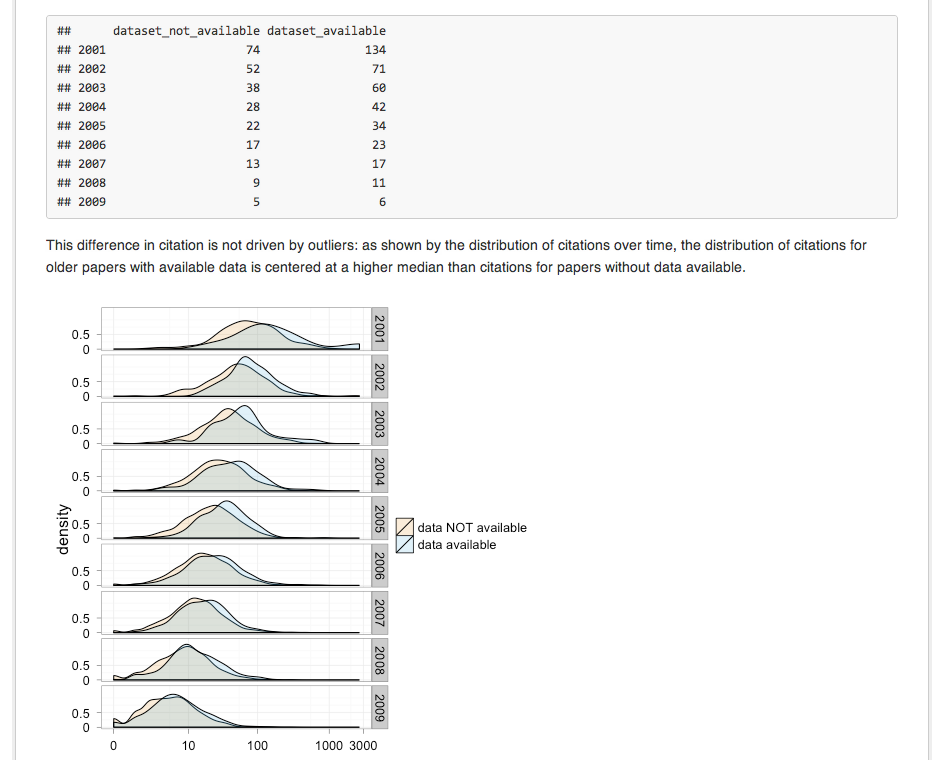
knitr + Markdown + GitHub = pre publication review
knitcitationscitet(c(Halpern2006 = "10.1111/j.1461-0248.2005.00827.x")) # then cite in your markdown file citet("Halpern2006")
# or read citations from a bibtex file which can be automatically generated and updated from services like Mendeley
bib <- read.bibtex("example.bib") # then cite inline citet(bib[["knitr"]])
knitcitations on Carl Boettiger's GitHub/
#