Thoughts on a preprint server
by Karthik Ram. Average Reading Time: almost 4 minutes.
In my last post I sang praises for markdown as a way to write and collaborate on manuscripts and other scientific documents. As easy as it is to use, the one command line step is enough of a barrier for most academics. This brought back an old idea that I batted around with a few folks right after ESA. With all the tools and web technologies that currently exist, it would be possible to create a pre-print server that runs on markdown and version controlled with git. Here’s a rough vision for how all the pieces might fit together.
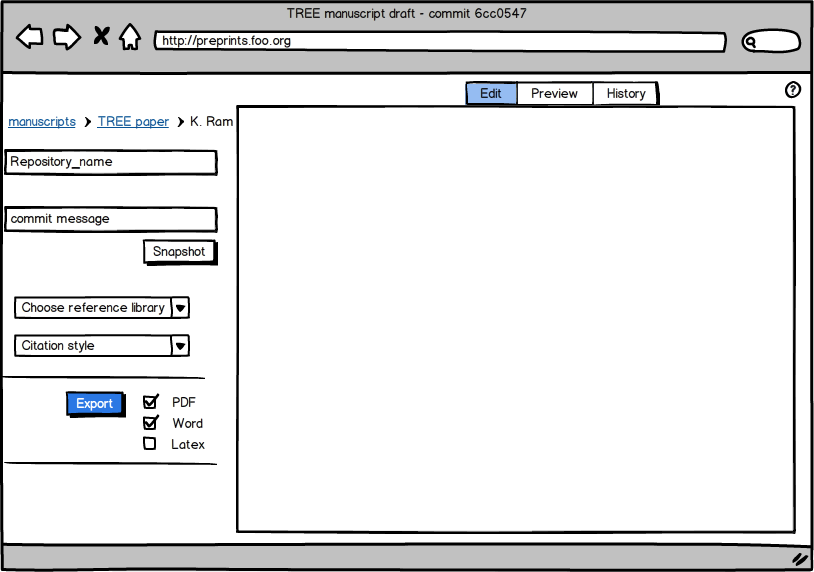
Creating a new manuscript
An author would begin by logging in with their GitHub (or similar) account. When a new manuscript is created, it automatically initializes a new git repository for the paper. Collaborators can be added using their GitHub accounts which automatically gives them write access. Documents are publicly readable by default (just like any GitHub repo) although one could change it to private if need be (which turns the repo private at the GitHub end).
Choosing a reference library
Next, authors choose a library to use with this manuscript. This could be either Mendeley or Zotero (or any other alternative) since both services have APIs and mechanisms for collaboration. With Mendeley, for example, it is trivial to read an up-to-date list of documents from a shared library using existing API methods. Both services also have bookmarklets which makes it easy to update missing references without ever leaving the browser (and all these efforts would show up on the desktop library at the next sync). An author could also manually upload a bib file. As authors type out citations in the editor window, the engine behind the web app autocompletes the process by reading from the JSON file (if reading from an API call).
Adding in tables and figures
Although it would be ideal to embed R/Python/other code directly into the MS and have knitr add in the results and figures, it’ll leave that out from version 1 of this hypothetical server. Instead, authors can just use an uploader and add in tables (as csv files), and figures (as images).
As the author builds up the manuscript, snapshots (git commits) can be saved at anytime with a human-friendly commit message. Even if an author doesn’t commit often, the document (and associated files) remain autosaved. At any time the manuscript can be previewed and exported into any format (with pandoc or other document conversion tool powering this part of the engine).
Authors familiar with git can skip the web app entirely and simply clone a copy, work locally, and commit back to the repo. This will appear seamlessly on the web version and remain transparent to co-authors.
Submitting the manuscript

When authors are ready to submit, it could be as simple as forking the repo over (although a little too soon for something this efficient). For now, one could export the final PDF, or if the journal has a write API, then submit directly via an API call and quickly fill out author info with a form. Ideally there would also be a link to the full repository so reviewers can see everything.
Some existing pieces
There are several pieces that could be hacked together to make a first draft of this work.
-
markdown preview – There are several implementations of live rendering a markdown preview to html. Here’s a particularly elegant one. Pandoc (or even Asciidoc) could also run behind the scenes and quickly parse the document.
-
Git bindings – Abstracting git from the user (avoiding issues like merging and merge conflicts for the time being) could be done using GitHub’s existing API.
-
Citations – This is already possible with the current version of Mendeley/Zotero API.
-
Stats – With all the rapid development on Shiny, executable papers with embedded R code aren’t far off. Here’s a neat prototype of a live, in-browser markdown file with embedded R code being parsed by knitr.
-
Comments – The issues feature on GitHub could be repurposed to serve as a feedback mechanism. Reviewers could refer to specific blocks of text using the line highlight feature.
Note: Although I mention a few services in the post (GitHub, Mendeley), the system is not dependent on these specific providers. Git repositories can be hosted anywhere (or even in multiple locations) and almost every reference manager can export citations as bibtex. GitHub just has the advantage of being a popular service (so a large user base), and already hosts the most number of academic papers, software, and code used in data analyses.

Some thoughts on building a preprint server. http://t.co/VHNsQoE8 cc/ @mfenner
Some thoughts on building a preprint server. http://t.co/VHNsQoE8 cc/ @mfenner
Fin idé här: ”Thoughts on a preprint server”. Versionshantering av manuscript i GitHub, integration med Mendeley – http://t.co/KPVAAzBG
We have a problem, or at least most publishers do. The problem is that most publishers are not in control of their core technology. I’m not in a position to be able to create an article submission API _yet_.
With a goal of getting a journal of the ground as quickly as possible, it makes sense to not rebuild the wheel, and in fairness to the company that runs eLife’s submission system, they have worked hard at creating the custom editorial flows that we wanted. However as there are so many publishers out there, with so many customisations required to the basic process of the editorial and peer review workflow, having an open API for submission is going to be a challenge.
What is the easiest thing that we could do? I think the easiest thing would be to think about how to make our initial lightweight submission become aware of external event triggers (like if your system creates a notification that some authors are interested in submitting.).
Right now I’m looking at how to build out a notification system that would work on publication to auto populate end points like Mendeley. I’ll put some thought into how that could be extended to work further up the processing stream.
@IanMulvany I wrote some more thoughts about how this might work.
http://t.co/VHNsQoE8
Some thoughts on building a preprint server. http://t.co/VHNsQoE8 cc/ @mfenner
Great set of posts on Markdown and publishing. I’ve been exploring the same issues outside of academia.
I’d be very interested in some more elaboration of your tables workflow. That’s the bit that I’m finding hardest to tackle.
Thanks for the note, Mike. I’ll write another post on my tables workflow.
[...] someone asked about tables in markdown in the comments section of an earlier post, I thought I’d elaborate a little more. Since the [...]